What is this challenge all about?
For decades, drug companies considered RNA too polar and solvent exposed to accept small molecules (SMs). It’s now clear that RNA contains structures that SMs can and do bind. In theory, any stage of the RNA life cycle could be targeted with small molecules (RNA synthesis, processing, splicing, translation, and protein interactions). The goal of RNA targeting mechanism is to inhibit or to increase the level generation of the protein.
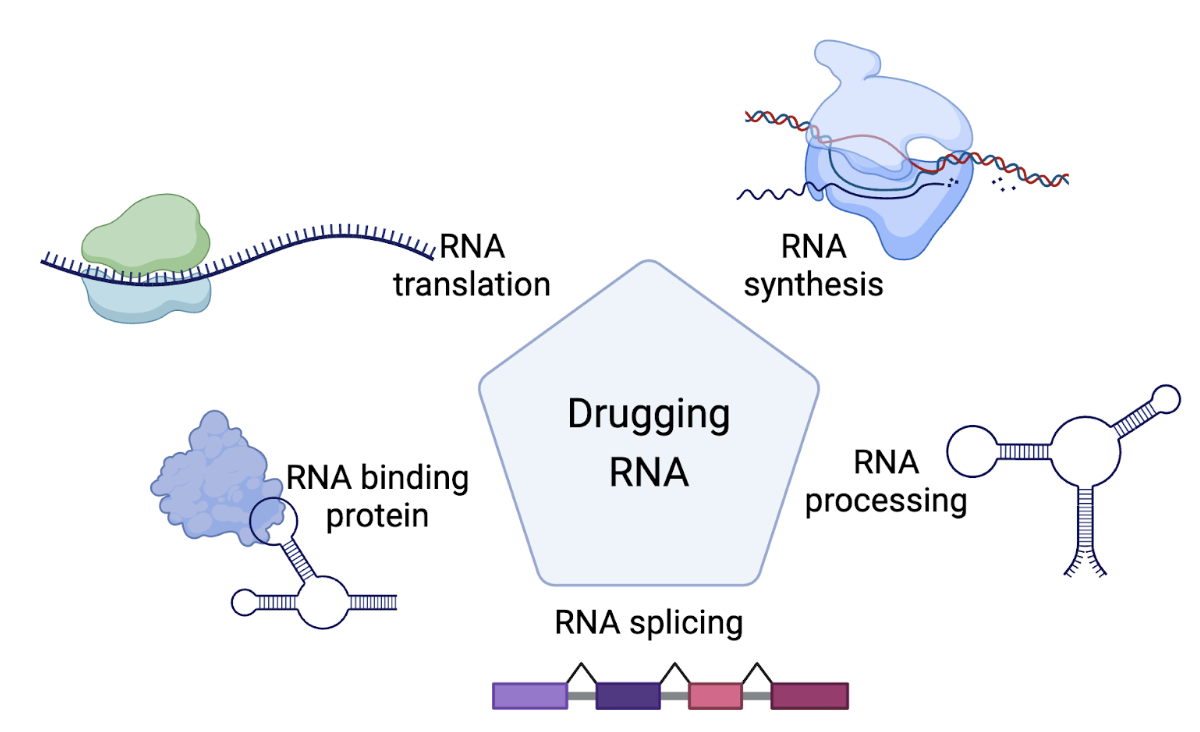
Modulating RNA biological function by RNA-targeting SMs can be extremely challenging yet its potential is huge, a successful RNA-targeting strategy could allow a systematic targeting of any protein or functional RNA previously considered undruggable at the protein level. The first and only SM FDA-approved drug targeting non-ribosomal RNA, risdiplam (Evrysdi), was approved for treatment of spinal muscular atrophy in August of 2020.
The field of RNA-targeting is just emerging and high throughput screens for small molecule discovery are still challenging. Development of RNA-specific computational tools is expected to help the rational design of RNA-targeting drugs, much as similar tools have helped protein targeted small-molecule drug discovery.
Problem statement
The goal of targeting RNA directly using small molecules is to increase or decrease the overall levels of RNA in a specific cell type or tissue of interest, thereby affecting protein expression levels. The process includes several major steps such as the selection of RNA targets as well as the regions within the RNA target to be targeted via small molecule screening. Identifying these regions and how small molecule binding may affect the RNA functionality is a key challenge today. Typically, RNA can be thousands of bases long and will dynamically fold in some local stable forms so identifying where to target the RNA to modulate translation, splicing or induce degradation of the RNA is challenging.
This process requires identifying an area to bind and a functional outcome. A secondary goal is to provide a 3D structure of the druggable pocket to target to allow for small molecule drug design.
Why is this problem important?
- By drugging RNA, we can effectively expand the druggable space:
– Only 1.5% of the genome that gets transcribed into RNA translates into proteins, hence there is a 98.5% opportunity - It will potentially allow to target the full spectrum of proteins with a single technology.
- It will allow us to target functional RNA (non-protein coding).
The existing approaches employed today
There are several approaches used by scientists today to identify regions in RNA to target using small molecules:
- Scanning the RNA target using secondary or 3D structure prediction algorithms (e.g. RNAfold). Predictions yielding RNA structures considered druggable (e.g. bulges or internal loops) can be good starting points for small molecule screening. Oftentimes, scientists have internal data linking predicted structures with small molecule moieties (e.g. Inforna) to help inform their selection of RNA targets/regions or this information may sit in publicly available datasets (R-BIND, ROBIN, HARIBOSS) and could be mined.
- Experimental mutational studies in functional RNA regions such as the UTRs, mRNAs splicing junctions or similar regions of functional importance in lncRNA. Identifying a particular mutation, deletion or insertion affecting RNA levels or its translation can be game changing. Today this is performed either retrospectively by looking at how genetic variation in cell types or human populations affect RNA/protein levels (e.g. by integrating GWAS, transcriptomics and proteomics datasets) or prospectively by defining an experimental system resembling the relevant disease context, setting up methods to introduce mutations in the lab and evaluate their functional effects.
- Experimental determination of RNA structure e.g. using probing methods such as SHAPE or high-resolution but low-throughput approaches such as NMR.
- Discovery of functional regions bound by RNA Binding Proteins (RBPs). This can be both experimental (e.g. CLIP) or data-driven (e.g. ENCODE dataset integration or predictive such as RBPmap).
A key limitation of the structure determination approaches (both experimental and prediction) is that it most often ignores the presence of that same structure elsewhere in the coding / non-coding genome, which may result in off-target effects. Once a small molecule that was screened to bind to RNA is characterized using biophysics, methods such as ChemCLIP are employed to investigate off-target effects.
The desired solution
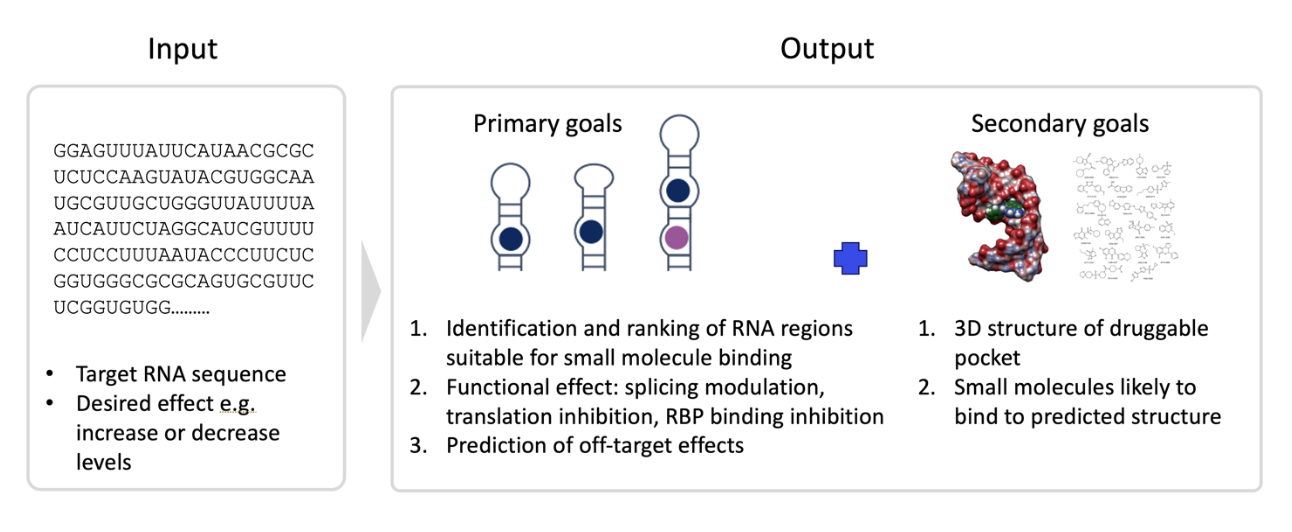
We envision a computational platform that takes as input an RNA target name or an RNA nucleotide sequence and a desired mechanism (e.g. increase or decrease RNA expression). The solution will provide as output:
- All the druggable sequences/structures along the RNA sequence that the pharma partner can use for small molecule screening (could be none) including:
– How often you can find same/similar sequences/structures across the coding / non-coding genome, which would suggest how likely we will see off-target effects
– How conserved is the sequence/structure across species e.g. monkey/mouse/rat
– How stable is the structure - Likely functional effect of small molecules binding to each of the structures e.g. affecting splicing, translation, inhibition of RBP binding, or otherwise just a good pocket for small molecules to bind
- A ranking / prioritization of the sequences/structures for likely hit finding success in a small molecule screen
- A prediction of which small molecules are likely to bind to the highly ranking sequence / structures from specific library e.g. commercial avaialble
- Other nice to haves:
– A 3D structural model of the highly ranking sequence / structure
Suggested proof of concept
- Partners select 1-2 PoC RNA targets (a high profile undruggable one) and the technology returns:
– Sequence / structures for small molecule screening
– Predictions of small molecules that are likely to bind - Partner then follows up by:
– Validating the structures that were predicted?
– Running a small molecule screen to identify small molecule binders
– Purchasing the small molecules that were predicted and test them in-house e.g. using biophysical assays
Further reading
- Targeting RNA structures with small molecules. Nature Reviews Drug Discovery volume 21, pages736–762 (2022).
- Drugging RNA. Nature Biotechnology volume 41, pages745–749 (2023).
- Computer-aided design of RNA-targeted small molecules: A growing need in drug discovery. Chem 7, 2965–2988 (2021)