Challenge
AI Powered Discovery to Enable Molecular Glue Therapies challenge

What is this challenge all about?
Proteolysis-targeting small molecules hijack the cellular protein degradation machinery to selectively reduce a target protein’s abundance. This new drug modality has the potential to degrade proteins that have been considered undruggable with existing approaches. The field is divided into two main types of molecules: PROTACs (PROteolysis Targeting Chimeras), which are molecules designed to bind a protein of interest (POI) to a chosen E3 ubiquitin ligase (E3); and molecular glues, which are small molecules designed to enhance a natural POI-E3 interaction. Molecular glues are easier to develop as oral drugs since they resemble small molecules that have been developed by pharma for many years.
There are over 600 human E3 proteins that differ in functionality across different tissues, cell types and conditions. This family of proteins control cellular processes by binding other proteins and tagging them for degradation. Once a specific POI is identified as naturally being tagged by an E3, we can design a small molecule drug to strengthen this interaction, thereby increasing the rate of degradation of the POI and modulating disease-causing activities. However, the main challenge for developing such molecules, named molecular glues, is the identification of naturally occurring POI-E3 interacting pairs.
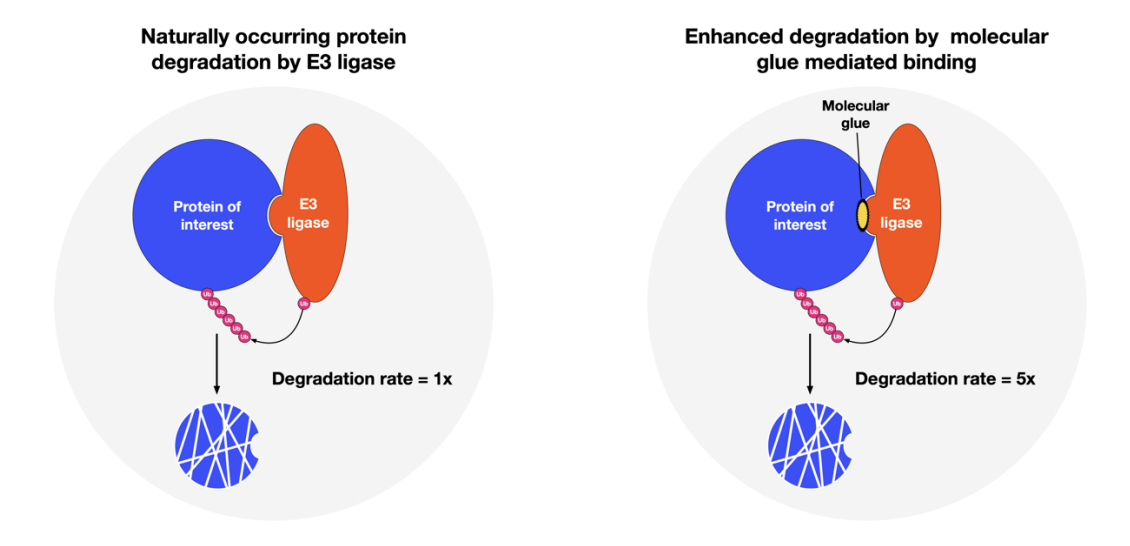
The following figure illustrates how molecular glues are expected to work. On the left, a naturally occurring process within a cell and on the right, enhanced degradation rate enabled by a molecular glue that strengthens the association to E3 ligase
We invite you to propose the development of a platform that enables molecular glue discoveries focusing on the following key challenges:
• An approach to identification of naturally occurring POI-E3 pairs in specific organs, cell types and conditions.
• For a given POI, identify and rank the E3 most likely to degrade it in the specified organ, cell type and condition.
• For a chosen E3, predict the proteins degraded in a specific organ, cell type and condition.
• For a selected pair of POI-E3, provide a 3D structure of the complex to enable the computational design of molecular glue.
How will you measure success?
The new technology developed by the company will be tested using the protein MYC as POI. There are multiple validated E3 for MYC in the scientific literature and the technology will be employed to test the predicted efficacy of specific E3 to degrade MYC in a variety of cell lines. We also expect to test a few novel E3 predicted to degrade MYC if such will be identified.
Can you provide an example that will demonstrate the impact of this technology and the complexity to address?
Small molecule drugs were successfully used to inhibit the function of enzymes by fitting into the functional pocket of the proteins. However, many proteins do not have a functional pocket in which a small molecule can bind with high affinity. MYC, used as an example above is such a protein. An additional example include KRAS. The ability to use a small molecule to degrade MYC or KRAS through an E3 will allow us to effectively target such proteins which are hallmarks of cancer. Here is one example that illustrates the challenges in targeting MYC with existing approaches.
There are several complexities within this challenge. First, showing the a protein can be tagged by E3 is not sufficient. The E3 activities vary across cell types, organs and conditions. In addition, one E3 can degrade many other proteins leading to potential side effects of drugs that employ a specific E3. Secondly, we do not have extensive data where E3 tagging of specific proteins have been documented. Data exists but it is limited and mostly public. Pharma companies do not have extensive data on this type of interaction. Finally, there are no clear and clear cut experimental validation systems that can be easily used. Often, validation is done by knocking out a specific E3 and measuring protein abundance changes within cell lines. This is an indirect type of measurement.